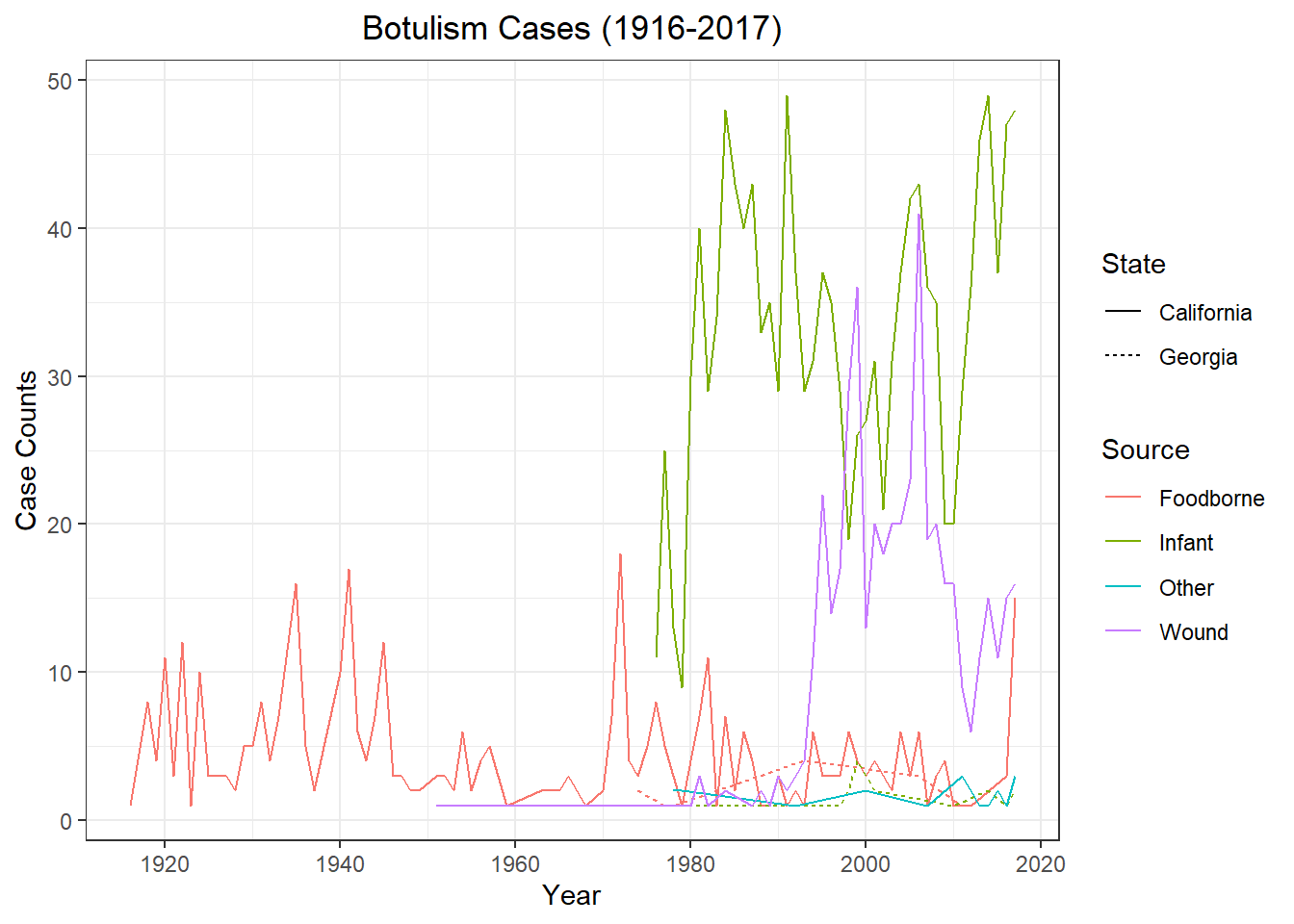
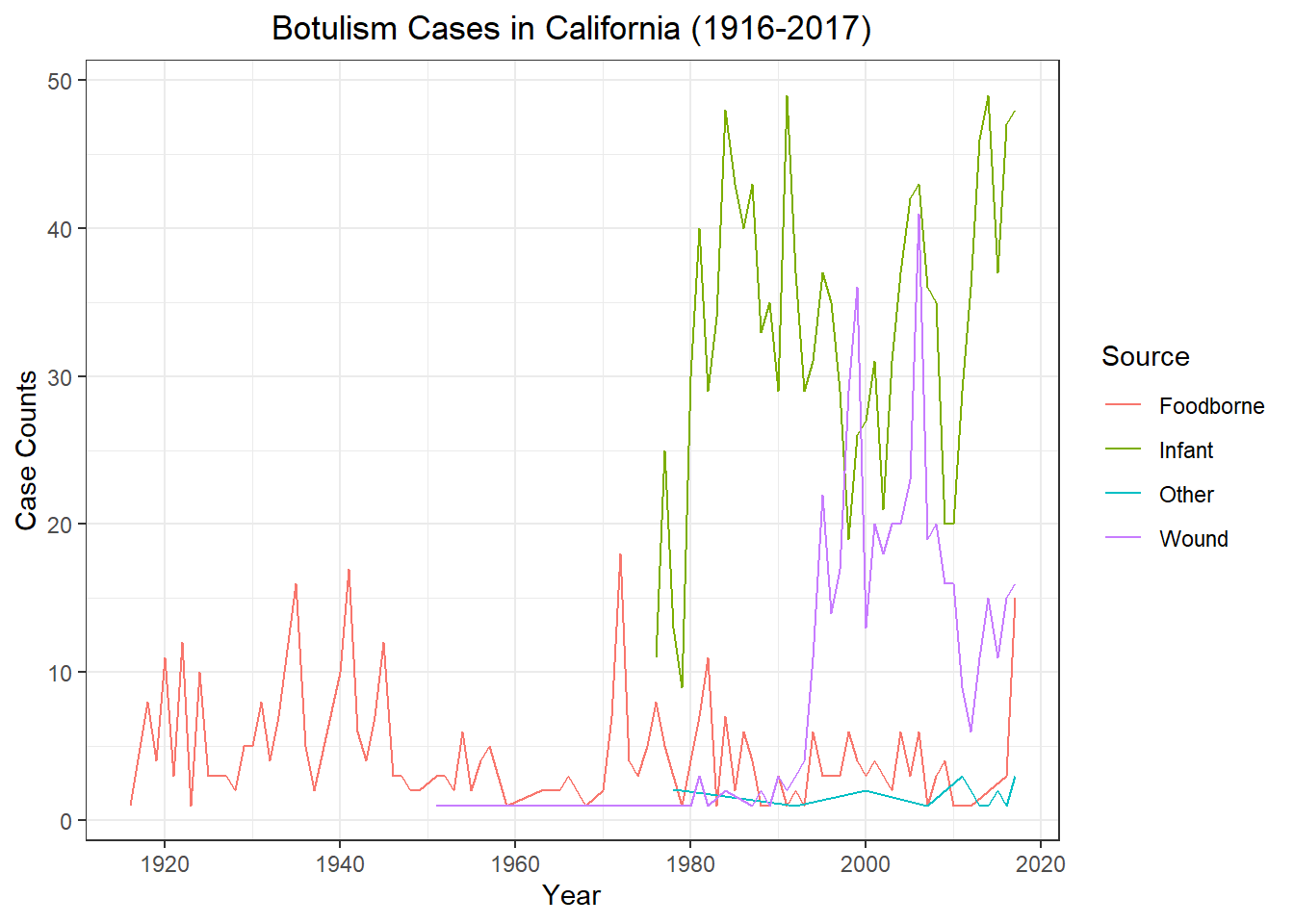
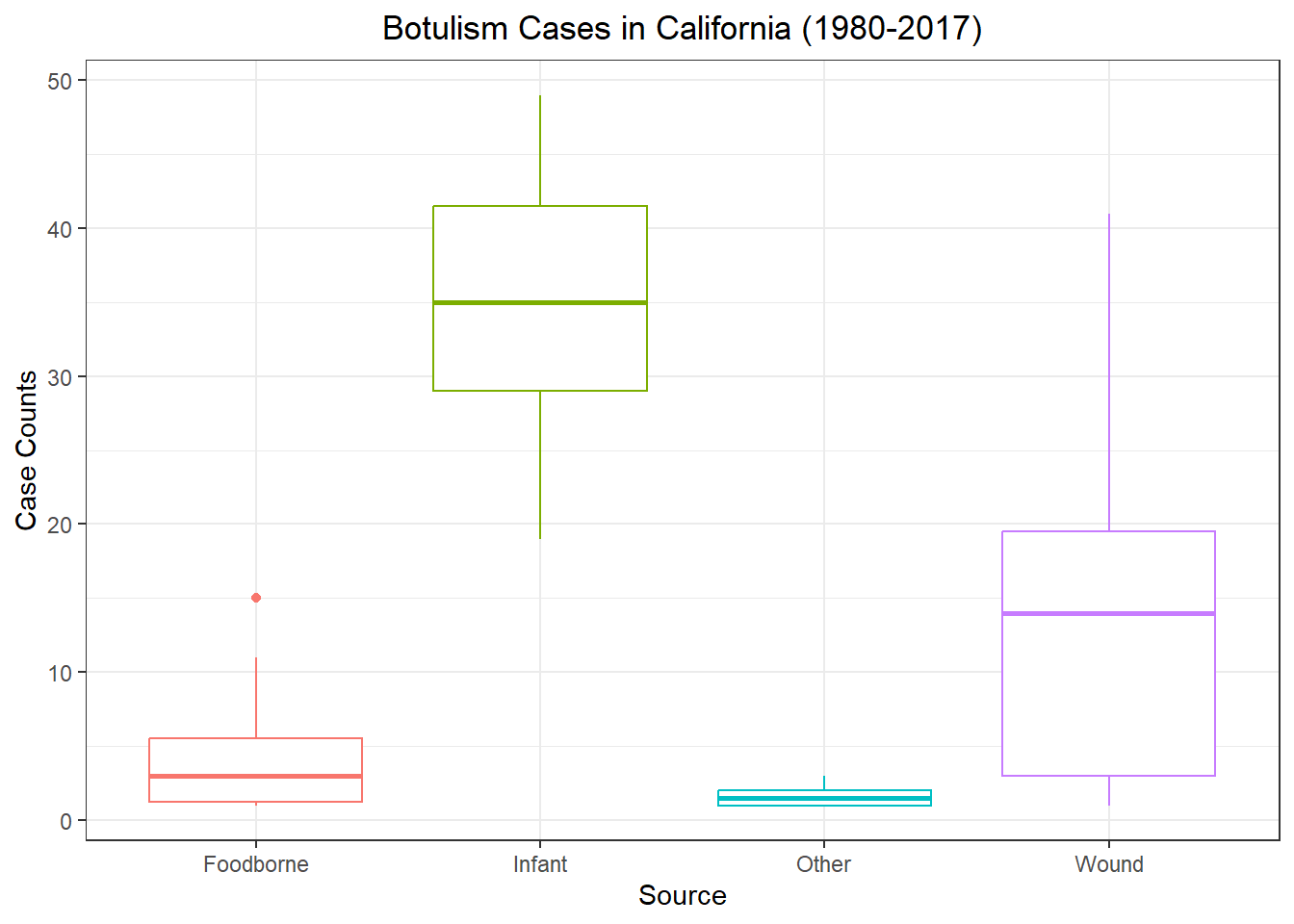
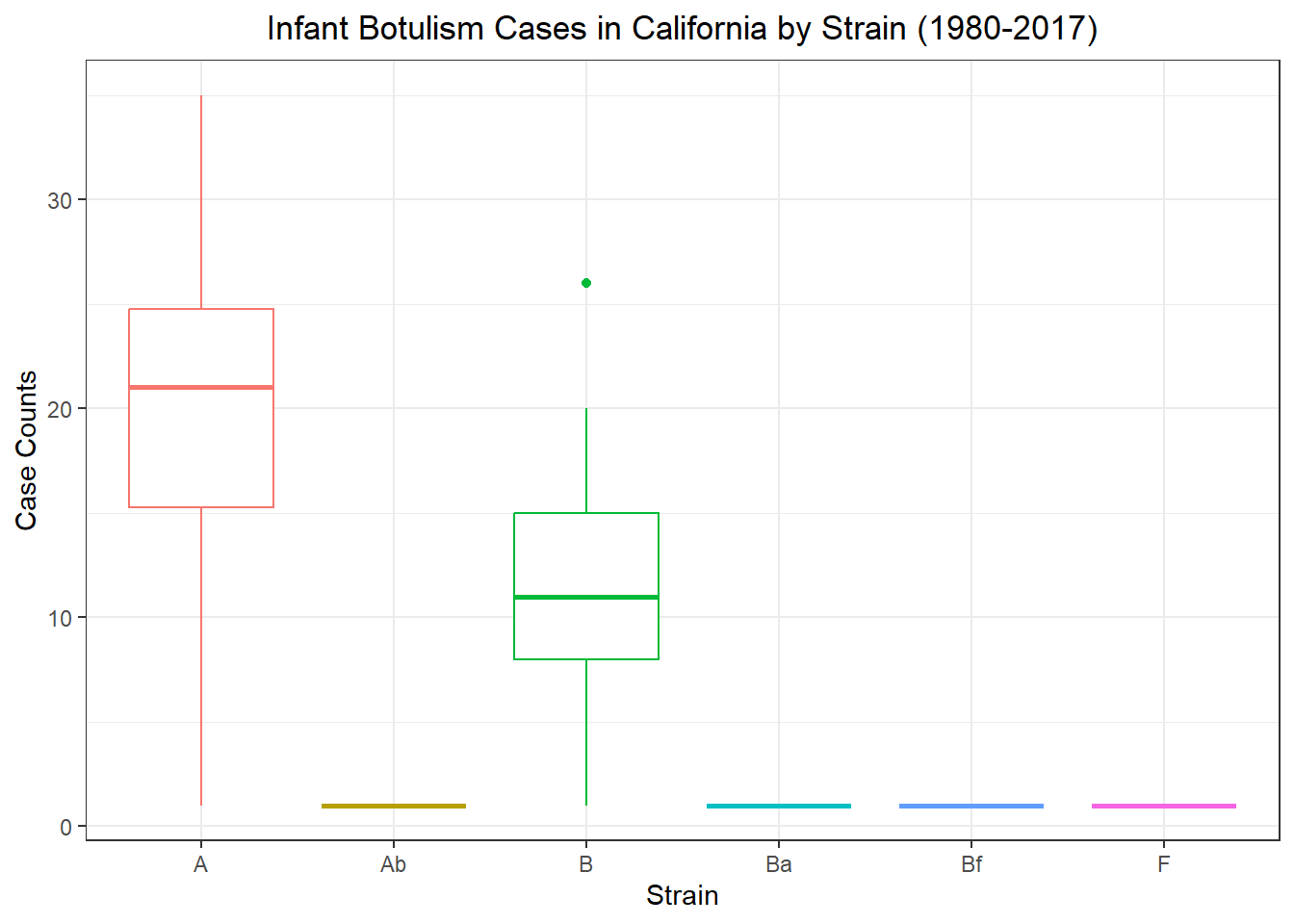
spc_tbl_ [2,280 × 5] (S3: spec_tbl_df/tbl_df/tbl/data.frame)
$ State : chr [1:2280] "Alaska" "Alaska" "Alaska" "Alaska" ...
$ Year : num [1:2280] 1947 1948 1950 1952 1956 ...
$ BotType : chr [1:2280] "Foodborne" "Foodborne" "Foodborne" "Foodborne" ...
$ ToxinType: chr [1:2280] "Unknown" "Unknown" "E" "E" ...
$ Count : num [1:2280] 3 4 5 1 5 10 2 1 1 1 ...
- attr(*, "spec")=
.. cols(
.. State = col_character(),
.. Year = col_double(),
.. BotType = col_character(),
.. ToxinType = col_character(),
.. Count = col_double()
.. )
- attr(*, "problems")=<externalptr>
State Year BotType ToxinType
Length:2280 Min. :1899 Length:2280 Length:2280
Class :character 1st Qu.:1976 Class :character Class :character
Mode :character Median :1993 Mode :character Mode :character
Mean :1986
3rd Qu.:2006
Max. :2017
Count
Min. : 1.000
1st Qu.: 1.000
Median : 1.000
Mean : 3.199
3rd Qu.: 3.000
Max. :59.000